Loop gains and losses between samples
This is a workflow on how to use AQuA-tools in a Docker or Tinker environment, and is designed to help you create meaningful subsets after calling loops.
We will use the following AQuA tools:
Motivation
In the previous recipe we called interactions directly from the contact matrix using extract_bedpe. This approach is great for finding loops, but figuring out which ones are biologically meaningful will require filtering. In this recipe, we introduce several AQuA-tools that help create meaningful subsets from genome-wide loop calls.
In this recipe, we’ll use two independent cell-lines:
sample1: H3K27ac HiChIP performed on the cell-line GM12878
sample2: H3K27ac HiChIP performed on the cell-line K562
Chapter 0: Ingredients
I. Prerequisites
You must complete the AQuA-tools Docker installation before starting, which can be found here. Alternatively, if you have access to Tinker, AQuA-tools are already pre-installed and no installation is needed.
II. Creating a workspace
This is an optional step. We encourage users to organize their analyses into a directory structure that keeps things nice and clean.
working_directory=/home/ubuntu/container_outputsmkdir -p $working_directorycd $working_directorymkdir -p \ data \ scripts \ results \ scratchIt’s nice to have all the files you want to play with in $working_directory/data
For now, we’ll follow the steps of this tutorial by housing ourselves in $working_directory/scratch, but we recommend writing .sh scripts $working_directory/scripts and storing the results in $working_directory/results
III. Gathering necessary files
For downstream analysis, we will need the following files:
- TAD
.bedfile - TSS
.bedfile - H3K27ac peaks
.bedfiles - Genome-wide loop
.bedpefiles
We’ll assign the .bed file paths to variable names for easier calling later:
cd $working_directory/scratch
# Initialise files in variablesTAD=/home/ubuntu/lab-data/hg38/reference/TAD_goldsorted_span_centromeres-removed_hg38.bed
TSS=/home/ubuntu/lab-data/hg38/reference/GENCODE_TSSs_hg38.bed
GM12878_peaks=/home/ubuntu/bed_files/GM12878_H3K27ac-peaks_ENCFF023LTU_hg38.bed
K562_peaks=/home/ubuntu/bed_files/K562_H3K27ac-peaks_ENCFF038DDS_hg38.bedFor the genome-wide loops .bedpe files, you must either run the last chapter in the previous recipe or have access to the pre-computed genome-wide loop files on Docker at /home/ubuntu/genome-wide_loops. If you are using the pre-computed loop files on Docker, we’ll rename them slightly for clarity in this recipe. If you computed the files yourself in the previous recipe, no re-naming is necessary.
# To use pre-computed genome-wide bedpe files on Docker
# navigate to scratch directorycd $working_directory/scratch
# GM12878# Make a copy in our scratch directorycp $HOME/genome-wide_loops/GM12878_H3K27ac_gw-loops_hg38.bedpe .
# Rename filemv GM12878_H3K27ac_gw-loops_hg38.bedpe GM12878_genome-wide-loops.bedpe
# K562# Make a copy in our scratch directorycp $HOME/genome-wide_loops/K562_H3K27ac_gw-loops_hg38.bedpe .
# Rename filemv K562_H3K27ac_gw-loops_hg38.bedpe K562_genome-wide-loops.bedpeIV. Defining samples
We’ll need a sample to get started. In Tinker or Docker environments, you’ll use the name of the sample as it appears on the Tinker box or Docker container.
# on Tinker or Dockersample1=GM12878_H3K27acsample2=K562_H3K27acNOTE: You can list all loaded samples on your tinkerbox or Docker container using list_samples
Chapter 1: Constraining interaction spaces
I. Intersecting peaks and TSSs
We can reduce the number of extracted loops to those that involve contact between TSSs and enhancers using intersect_bedpe, but first we want to make sure our TSSs are epigenomically active. To do this, we’ll use bedtools intersect to find TSSs that intersect with regions in our peaks files.
# GM12878bedtools intersect -a $TSS -b $GM12878_peaks > GM12878_active-tss.bed
# K562bedtools intersect -a $TSS -b $K562_peaks > K562_active-tss.bedII. Finding enh-tss loops
Next, we’ll run a two-bed intersect_bedpe analysis to identify loops with an active TSS or enhancer on either anchor. We’ll run intersect_bedpe two times, once for each sample’s genome-wide loops. To focus only on loops that connect an active TSS to an enhancer, we’ll filter the intersect_bedpe output for intersection types A-B or B-A. To learn more about intersection types, see our Docs.
# intersections for GM12878intersect_bedpe \ --bed_A GM12878_active-tss.bed \ --bed_B $GM12878_peaks \ --bedpe GM12878_genome-wide-loops.bedpe | \awk '$NF=="A-B" || $NF=="B-A"' > GM12878_gw-loops_tss-enh.bedpe
# intersections for K562intersect_bedpe \ --bed_A K562_active-tss.bed \ --bed_B $K562_peaks \ --bedpe K562_genome-wide-loops.bedpe | \awk '$NF=="A-B" || $NF=="B-A"' > K562_gw-loops_tss-enh.bedpeChapter 2: Merging interactions
I. Merge the called genome-wide loops into one file
Now that we’ve narrowed our loops to those that bring an active TSS and enhancer into contact, we will use union_bedpe to merge the spaces into a single .bedpe file. Here, we take the union of overlapping interaction regions from the two samples and create a unified set of candidate loops for comparison. For more information about how union_bedpe works, see our Docs.
union_bedpe \ --bedpe GM12878_gw-loops_tss-enh.bedpe \ --bedpe K562_gw-loops_tss-enh.bedpe > union_scaffold.bedpeChapter 3: Annotating interactions
I. Annotate union scaffold bedpe with contact values
Next, we will run a two-sample query_bedpe analysis to annotate each loop in the union scaffold with contact values and delta contact values. We’ll use --inherent TRUE to apply inherent normalization, which will standardize the contact values and allow us to compare interaction strengths between samples. To know more about inherent scores, check out our Docs.
In addition to retrieving contact strengths, query_bedpe can also refine our loop coordinates. By setting --formula max and --fix false, we update each loop’s coordinates to the bin pair with the highest contact value within the original region, allowing us to focus on the most relevant signal. For more information about how query_bedpe works, see our Docs.
query_bedpe \ --bedpe union_scaffold.bedpe \ --sample1 $sample1 \ --sample2 $sample2 \ --genome hg38 \ --formula max \ --fix FALSE \ --inherent TRUE > union_scaffold_inh-annotated.bedpeII. Threshold with standard inherent score 1
Since we normalized our contact scores with inherent normalization, we can easily select a threshold to filter our pairs. Most normalized loop scores fall between 0 and 1, with higher values indicating strong contact. When we subtract one sample’s normalized score from another to calculate delta, the resulting values are centered around zero. Here, we use a threshold of >1 to define loops with strong gains in contact, and <−1 to define loops with strong losses.
awk '$NF>= 1{print $0}' union_scaffold_inh-annotated.bedpe > loop-gains.bedpeawk '$NF<=-1{print $0}' union_scaffold_inh-annotated.bedpe > loop-losses.bedpeChapter 4: Visualizing interactions
I. Visualise gains and losses with plot_APA
We’ll use plot_APA to visualize the aggregated contact signal for our gains and losses. Since the number of rows differs between the two files, we’ll use --loop_norm TRUE to standardize the contact scores by how many loops are in the file. For more information about how plot_APA works, see our Docs.
plot_APA \ --bedpe loop-gains.bedpe \ --sample1 $sample1 \ --sample2 $sample2 \ --genome hg38 \ --loop_norm TRUE \ --out-dir ~/container_outputs/gains
plot_APA \ --bedpe loop-losses.bedpe \ --sample1 $sample1 \ --sample2 $sample2 \ --genome hg38 \ --loop_norm TRUE \ --out-dir ~/container_outputs/lossesHere’s our output!
Gains:
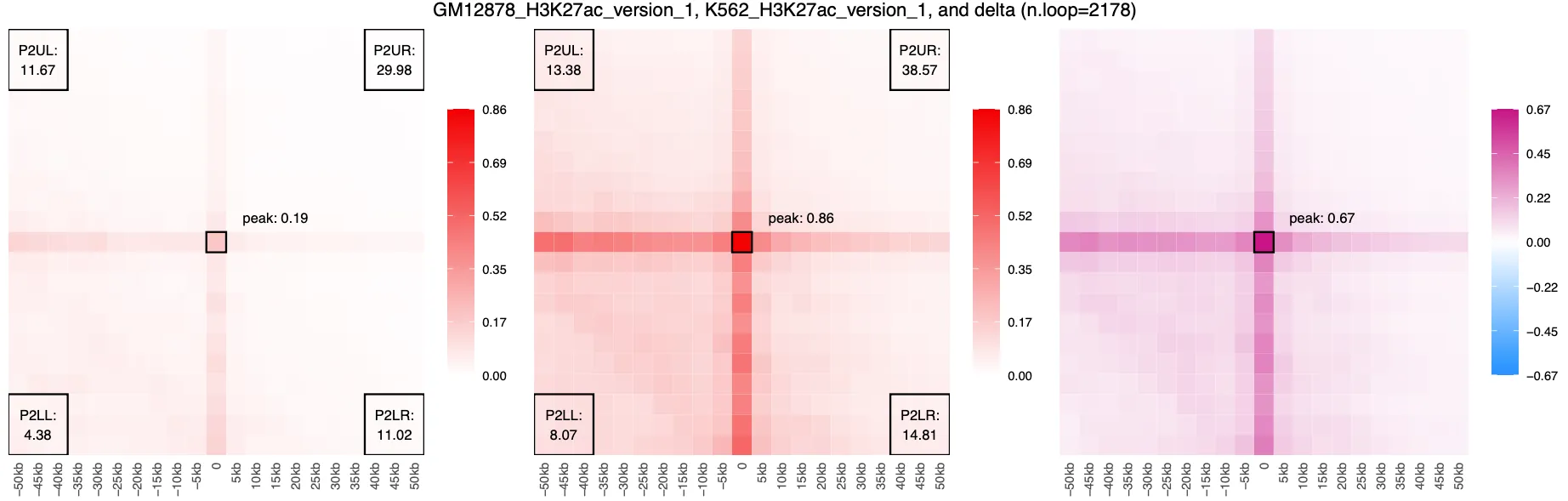
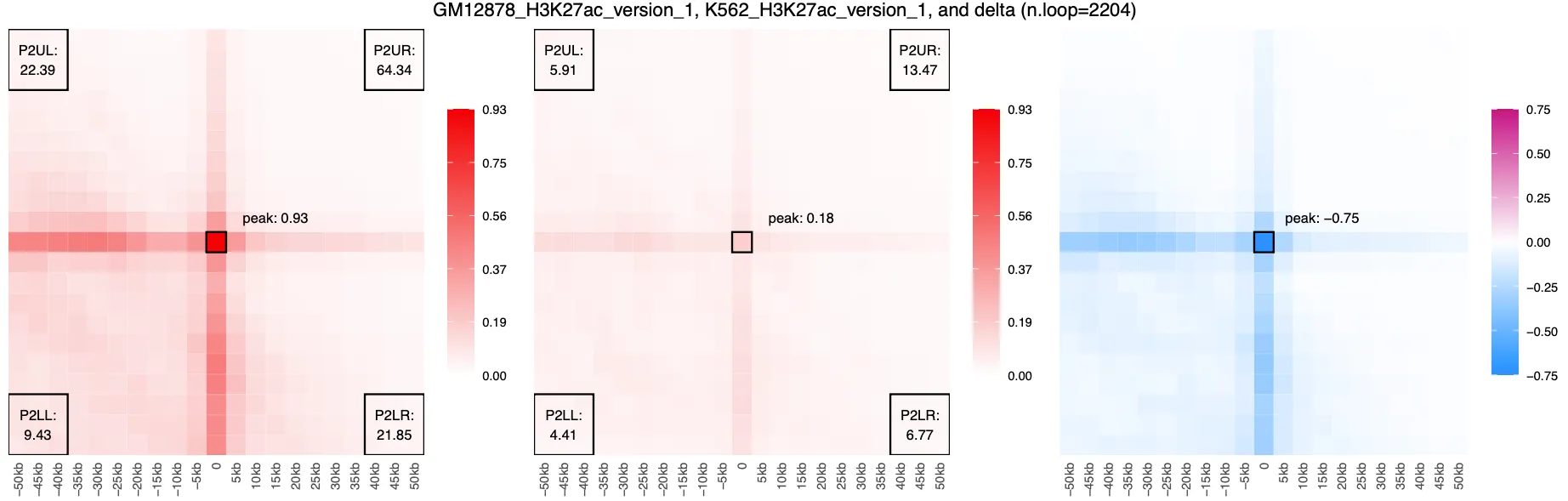
Losses:
Conclusion
By viewing the delta APA plots for loop gains and losses, we can visually confirm that our filtered subsets represent cell-type-specific contact enrichment. The magenta signal in the gains set indicates increased contact frequency in K562 relative to GM12878, while the blue signal in the losses set reflects loops with stronger contact in GM12878. These contrasting patterns suggest that the differential loops we identified are not random, but instead reflect meaningful biological differences in chromatin organization between the two cell types.
We created these loop subsets with a streamlined AQuA-tools workflow. We started with genome-wide loops called with extract_bedpe and used intersect_bedpe to find loops that bring active TSSs and enhancers into contact. Then, we used query_bedpe to refine our coordinates and annotate our loops with inherent normalized contact scores directly from the contact matrices. This allowed us to categorize loops into gains and losses based on a consistent threshold. From there, we used union_bedpe to merge loops from both cell types into a single scaffold for comparison. Finally, we visualized loop behavior between samples with plot_APA.
Our goal in this workflow was to highlight the power and flexibility of AQuA-tools for HiChIP analysis and demonstrate how this modular set of tools can guide discovery in complex 3D genomic data. With these types of workflows, we hope to support researchers in asking their own questions of the data without needing to build custom pipelines from the ground up.
In the next recipe, we’ll continue our exploration by looking into how to organize overlapping interactions into meaningful groups using cluster_bedpe. We’ll introduce annotate_cluster, which adds biological context to each cluster by incorporating peak files and gene annotations.