build_bedpe
Concepts
build_bedpe is often the first AQuA tool used in a 3D genomics pipeline. build_bedpe takes bed files as input, builds pairs between the elements in the bed files, and outputs a bedpe file. The input bed files can be the same file, or they can be different bed files. The following series of slides will use faux data and the four modes of representation to explain and visualize different use-cases and parameters for build_bedpe.
Homogeneous/self builds
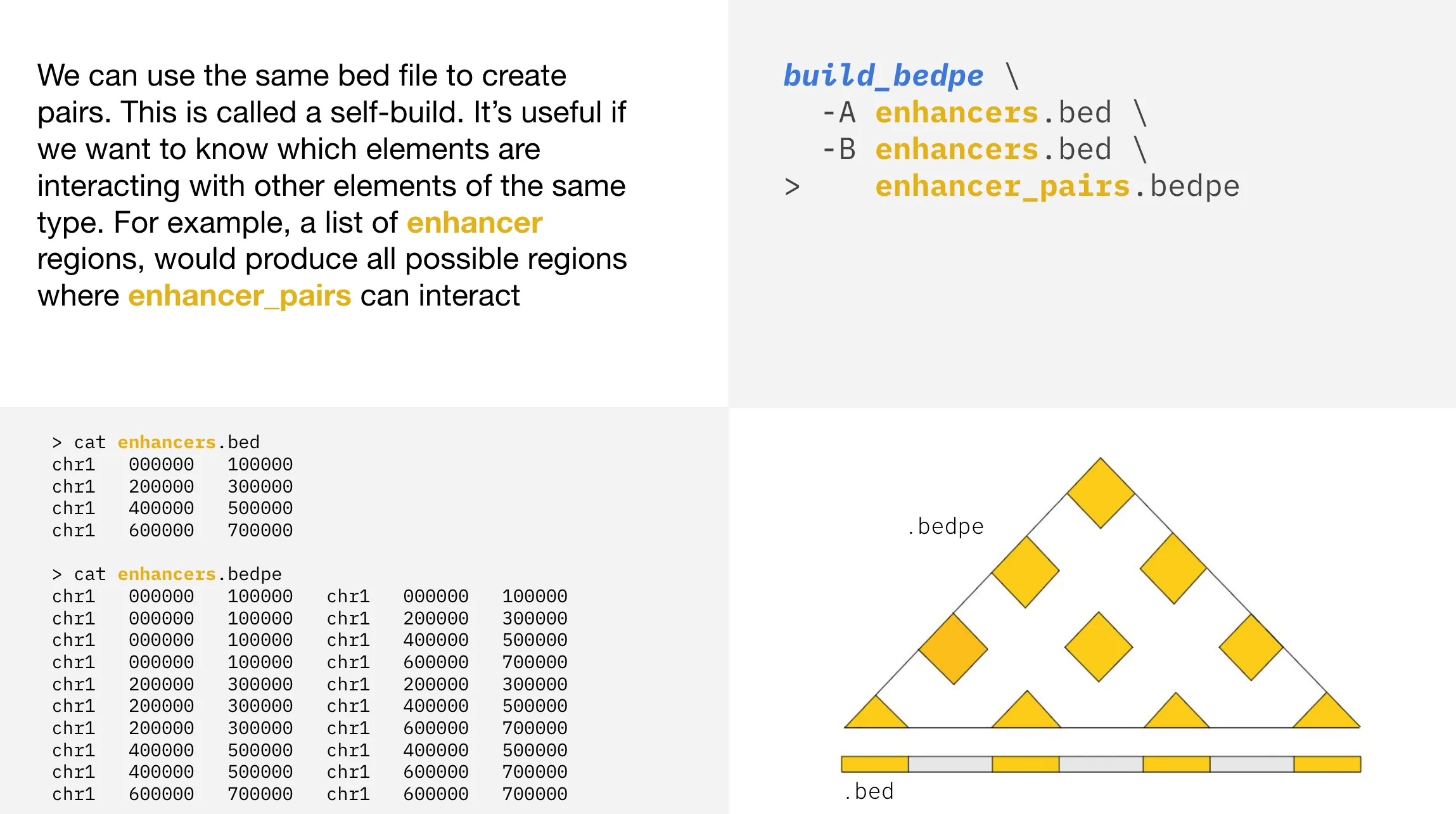
If there are four enhancers in enhancers.bed, a self build would result in 10 interactions. These interactions can be visualized as small yellow triangles inside a larger triangular plot. At the base of the larger triangle are four small yellow triangles representing the four enhancers in the bed file. Above these four enhancers would be three yellow triangles, representing the interactions between neighboring enhancers (enhancers 1 and 2, enhancers 2 and 3, and enhancers 3 and 4). The next higher level would show two yellow triangles, representing the interactions between every other enhancer (enhancers 1 and 3, and enhancers 2 and 4). The top level, at the tip of the triangle, would show one yellow triangle, representing the interaction between the most distant enhancers (enhancers 1 and 4).
Minimum distance constraint
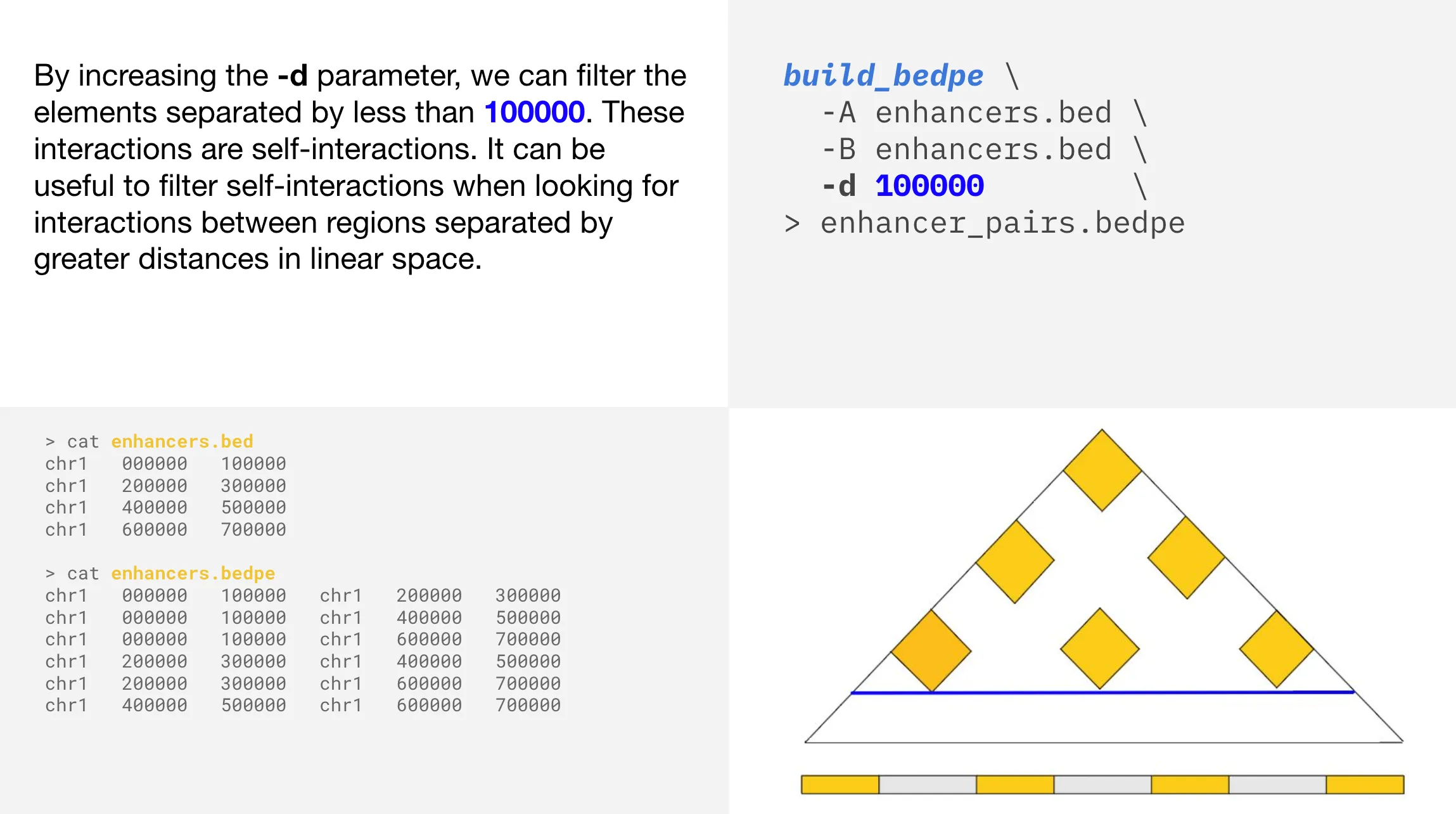
In this example, applying a -d parameter of 100000 removes the bottom four yellow triangles from the larger triangular plot. There are six remaining interactions in the plot, all separated by more than 100000 base pairs in linear space.
Maximum distance constraint
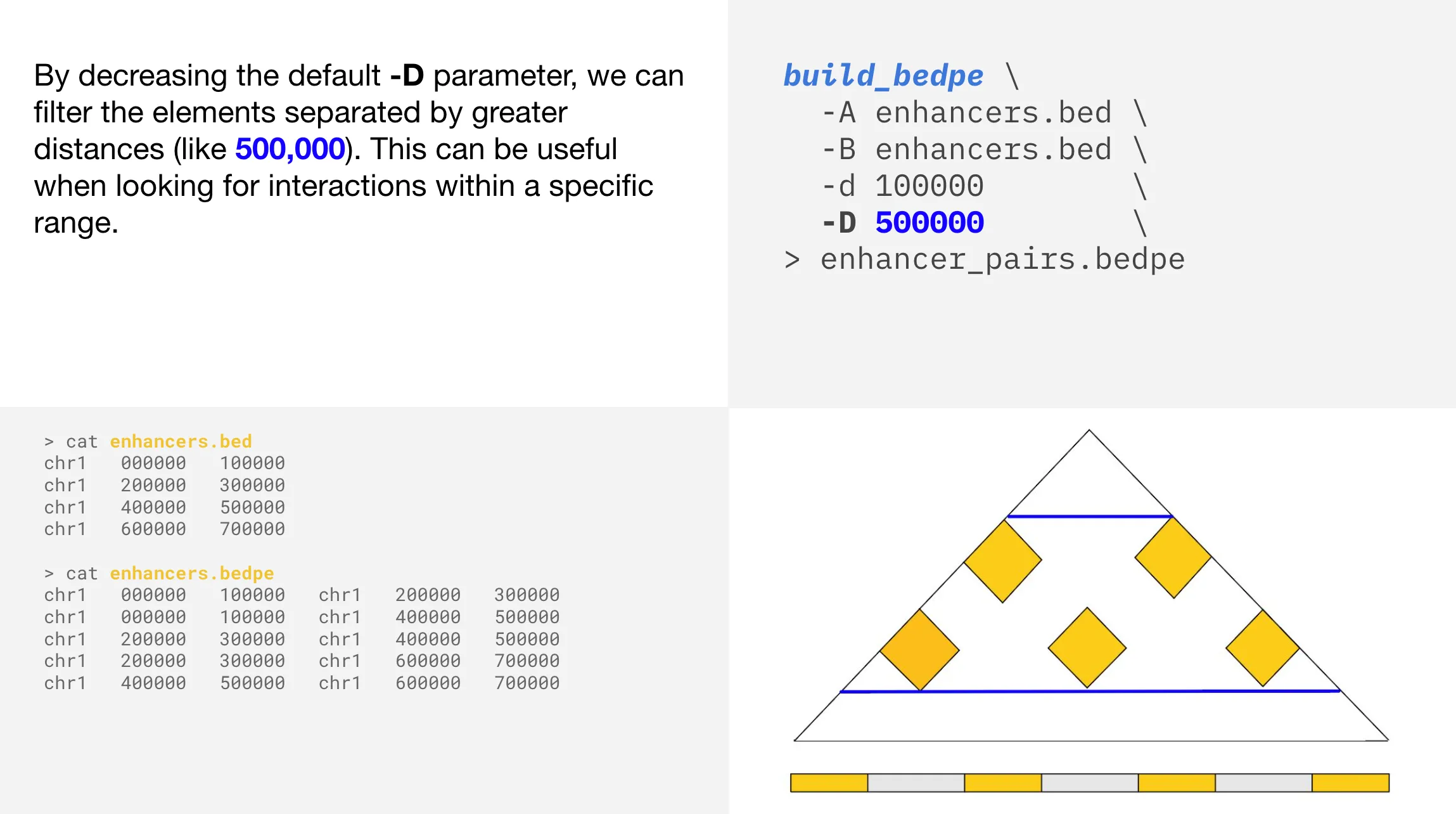
By applying a -D parameter of 500000 , we have now also removed the top yellow interaction triangle, which represents the interaction between the most distant enhancers in linear space. Visually, we are left with five mid-range interactions in the plot.
TAD distance constraint
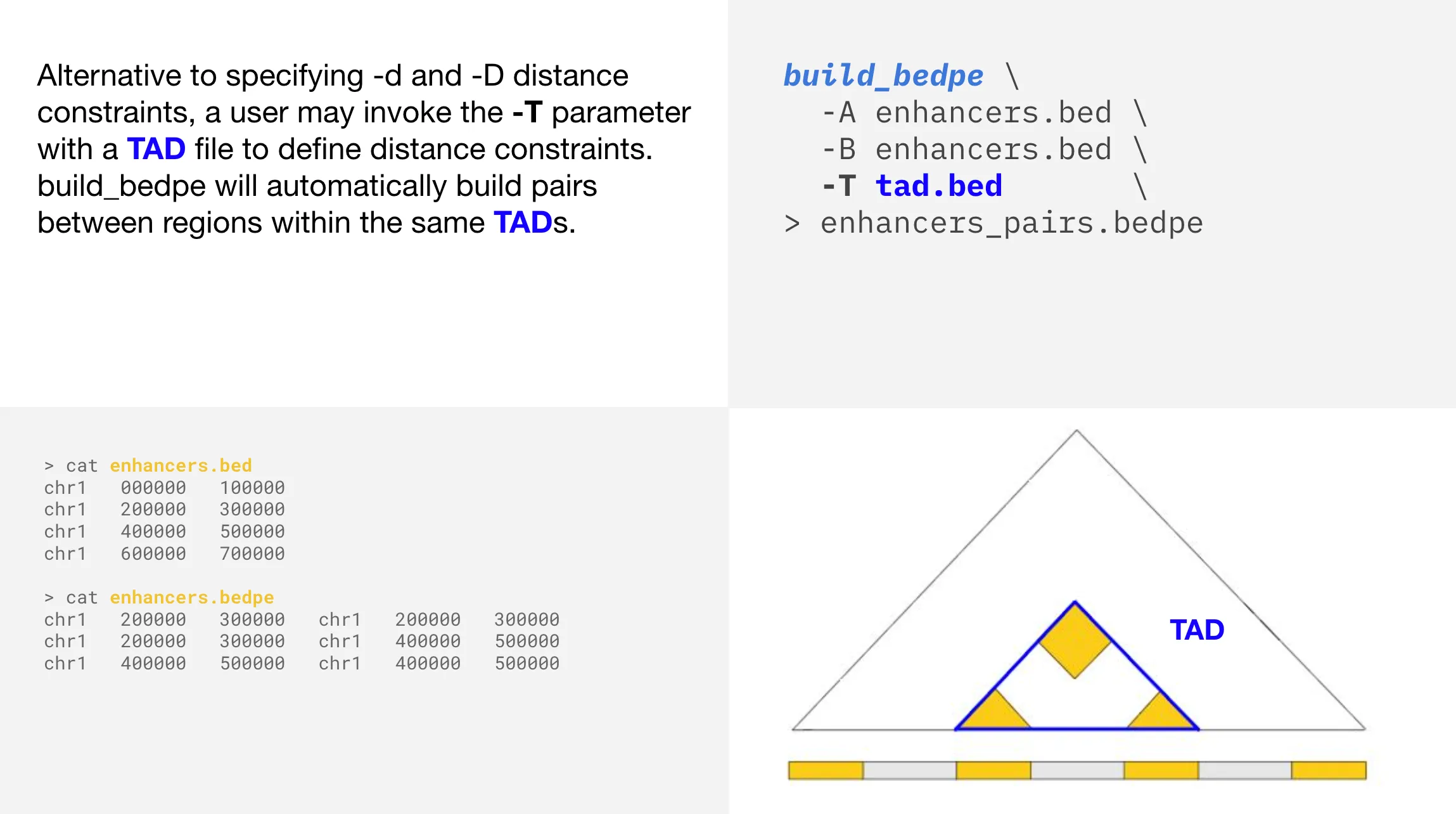
Heterogeneous builds
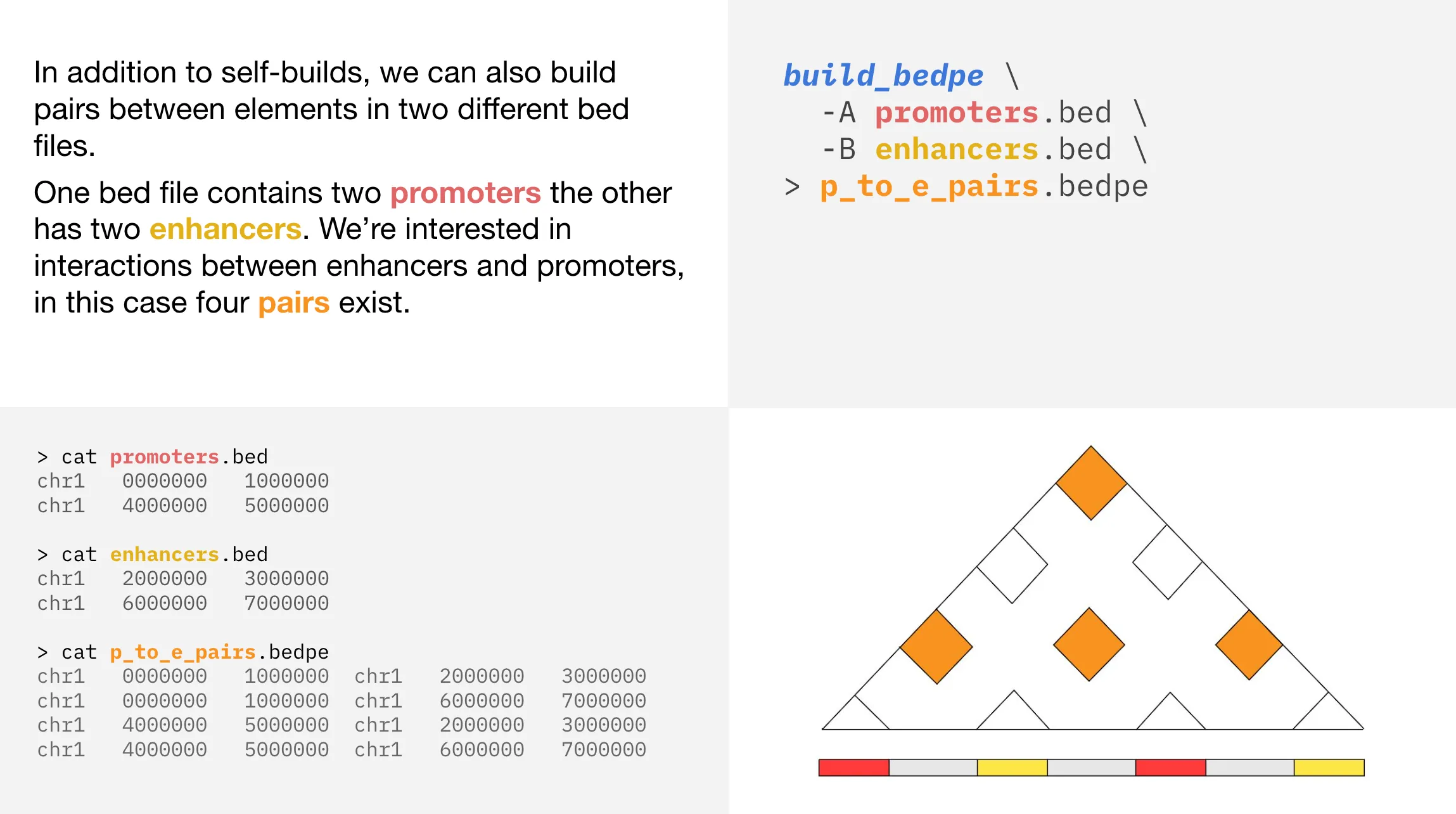
In this example, one bed file contains two promoters the other bed file has two enhancers. If we visualize the two promoters as red regions and the two enhancers as yellow regions, then we can use orange regions to represent the interactions between promoters and enhancers. In this case, there are four interactions represented by orange squares in the triangle plot. These interactions take place between enhancer 1 and promoter 1, enhancer 1 and promoter 2, enhancer 2 and promoter 1, enhancer 2 and promoter 2.
Usage
build_bedpe builds pairs between elements in two bed files. Pairs can be constrained by a third bed file (usually TADs) or by user-defined minimum and maximum distances. Pairs are printed in bedpe format to standard out. Pairs can be use to query .hic files with query_bedpe.
Usage and Option Summary
build_bedpe -A path/to/bed1.bed -B path/to/bed2.bed -T path/to/TADfile.bed(or):
build_bedpe -A path/to/bed1.bed -B path/to/bed2.bed -d 10000 -D 100000Required
| Short Option | Long Option | Description |
|---|---|---|
-A | --bed_A | Path to the first bed file |
-B | --bed_B | Path to the second bed file |
Optional
| Short Option | Long Option | Description |
|---|---|---|
-T | --TAD | Path of the TAD file to restrict pairings outside of the TAD |
-d | --min_dist | Minimum distance between pairs used to drop results. Default 0 bp |
-D | --max_dist | Maximum distance between pairs used to drop results. Default 5 Mb |
-i | --get_trans | If pairs between different chromosomes should be made. If TRUE, will print trans pairs only. Default FALSE |
-h | --help | Help message |