cluster_bedpe
Concepts
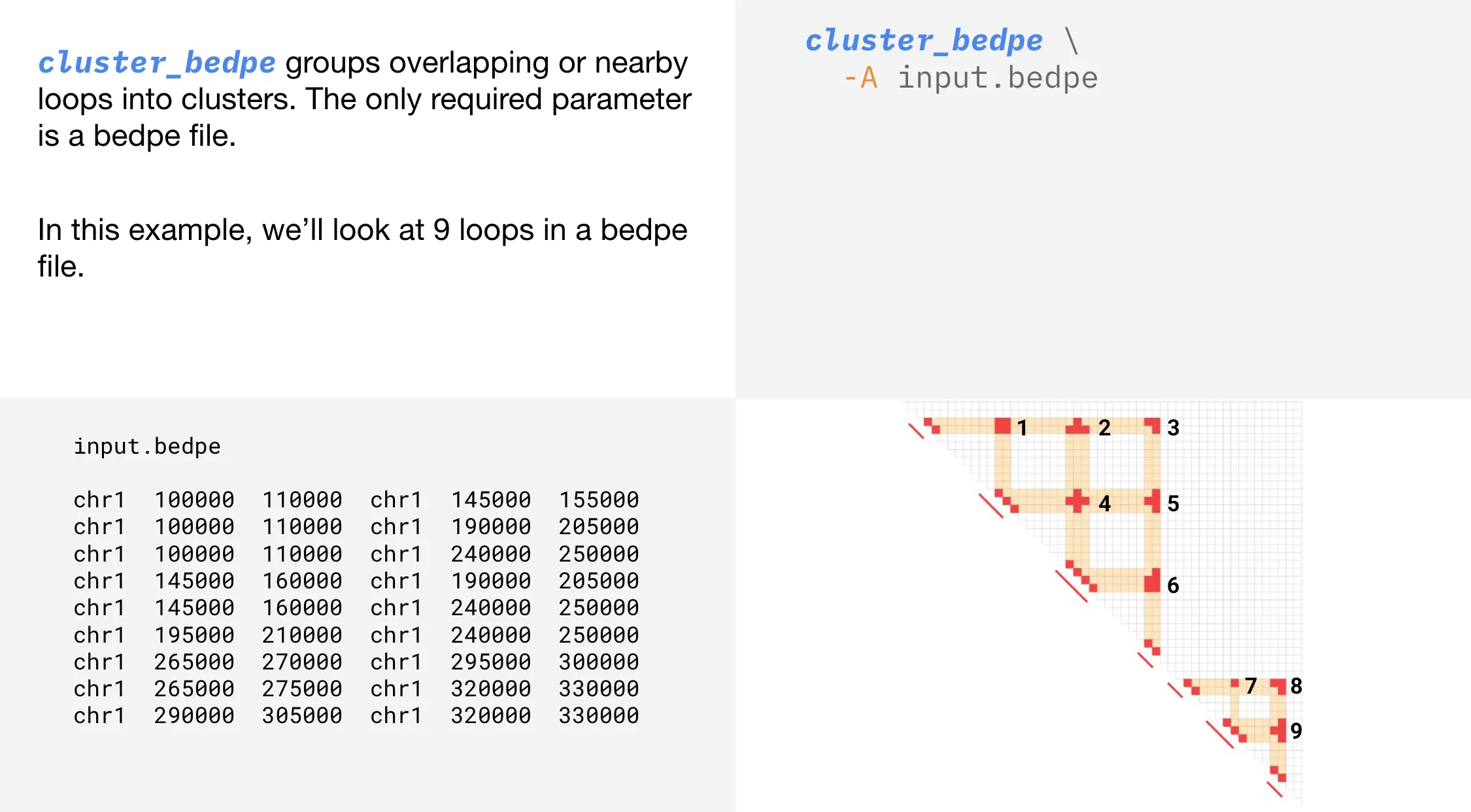
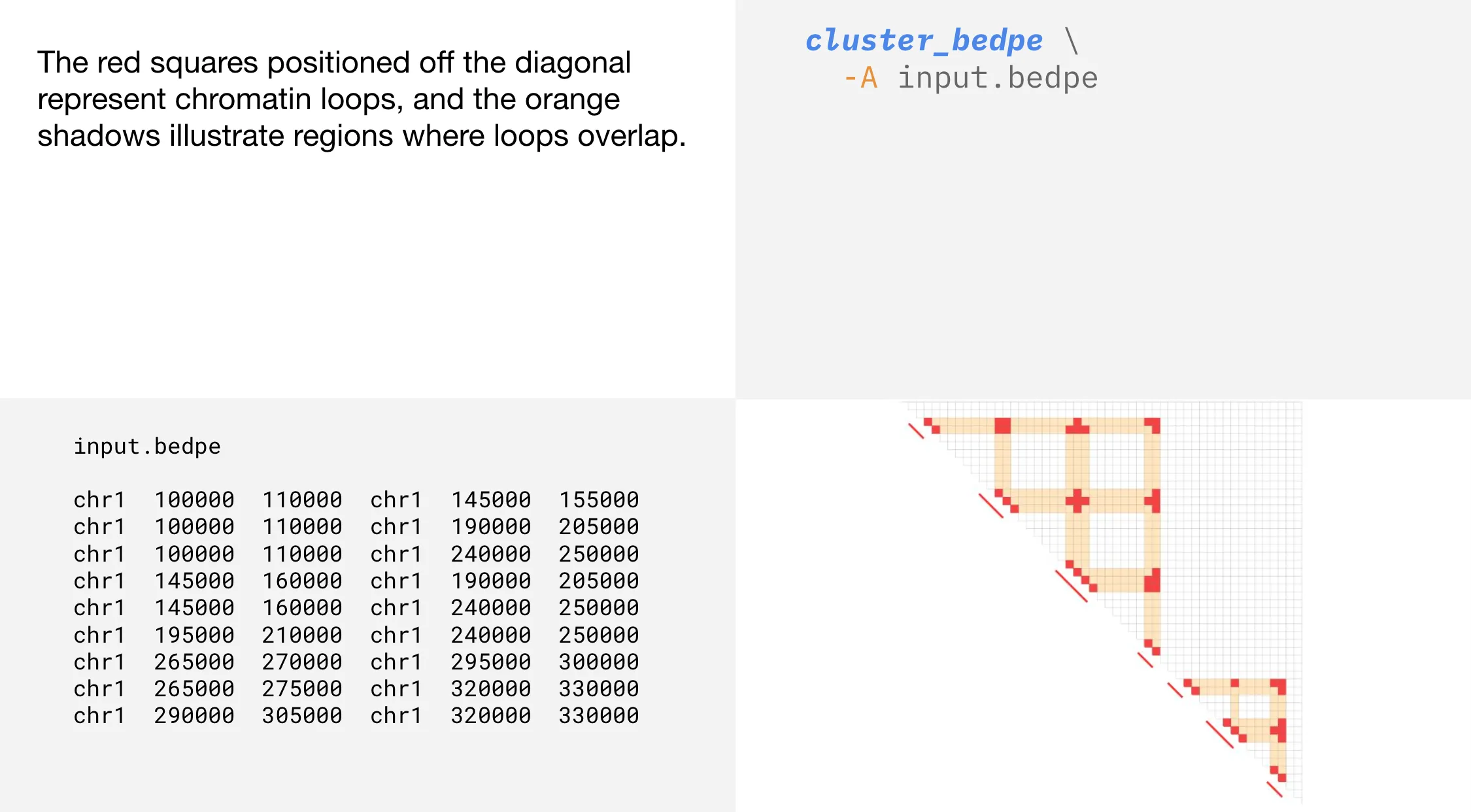
A loop is a connection between two regions in the linear genome that are brought close together in 3D space. Overlapping interactions occur when loops share anchoring regions on the diagonal.
cluster_bedpe groups overlapping interactions. Creating clusters from overlapping interactions provides a flexible, context-specific view of genome organization, capturing dynamic regulatory relationships that TADs may miss.
cluster_bedpe is often used after extracting loops from a .hic file with extract_bedpe.
Input
Output

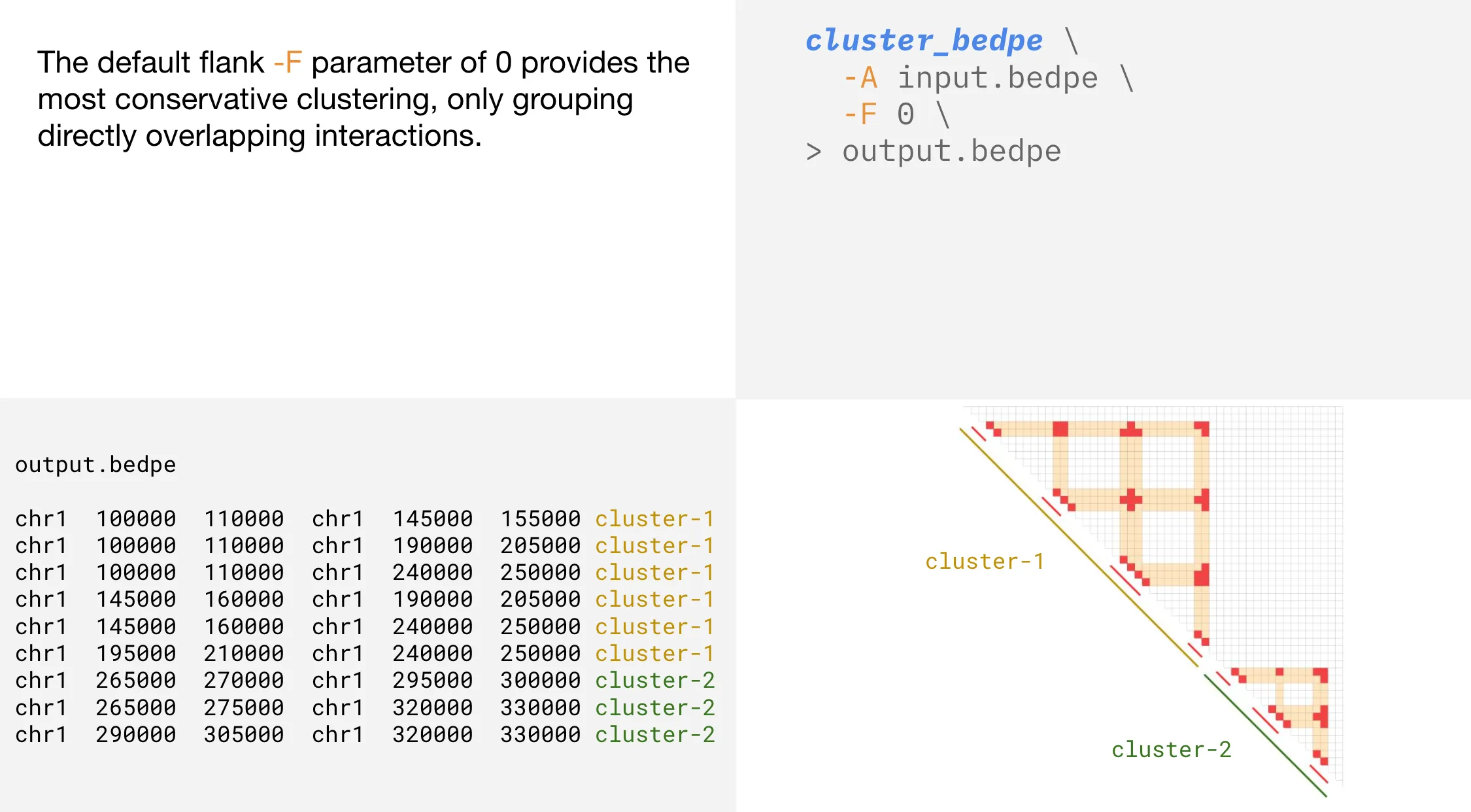
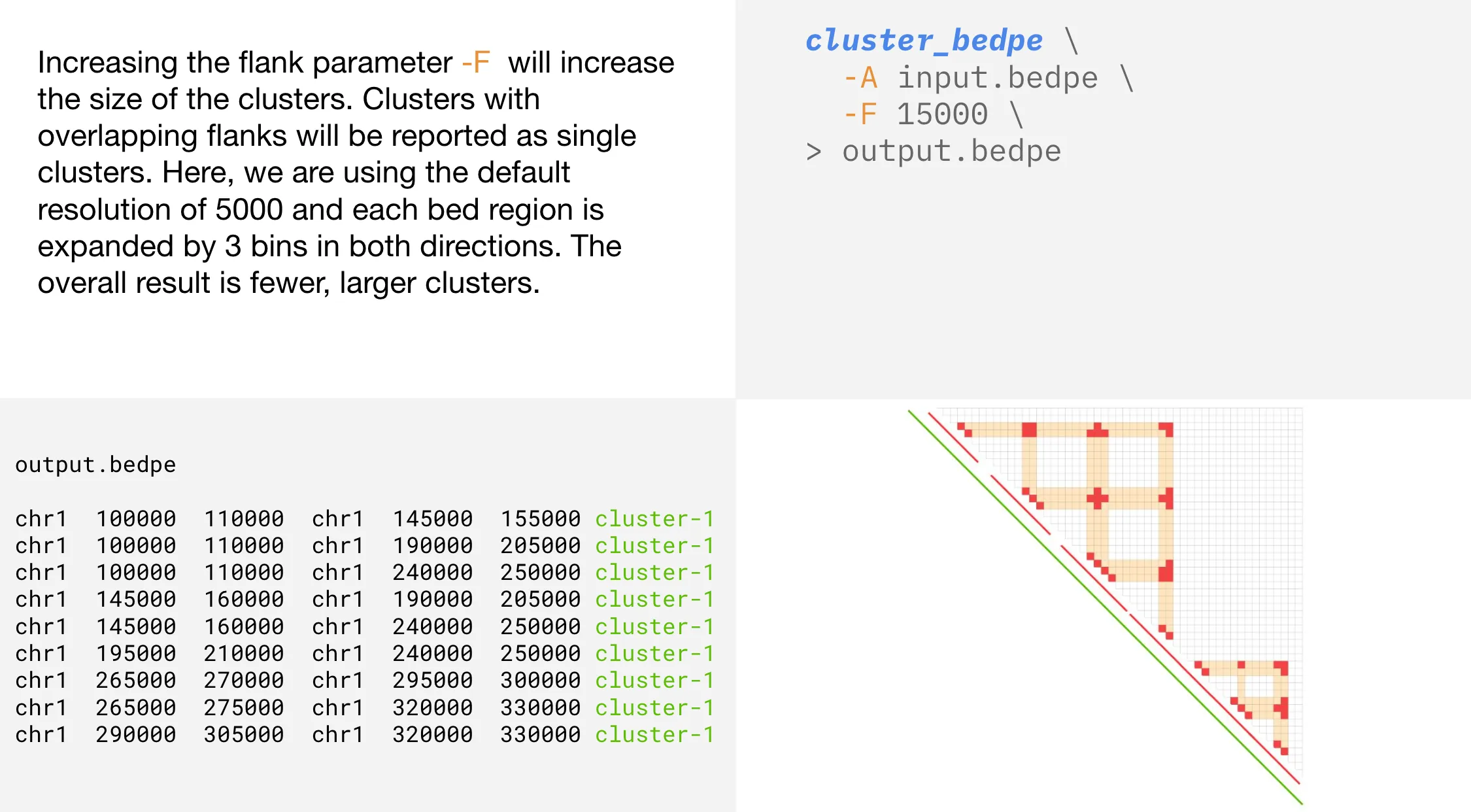
Flank
Usage
cluster_bedpe clusters bedpe rows based on basepair intersections
Usage and Option Summary
cluster_bedpe -P file.bedpeRequired
| Short Option | Long Option | Description |
|---|---|---|
-P | --bedpe | Path of the bedpe file you want to use |
Optional
| Short Option | Long Option | Description |
|---|---|---|
-F | --flank | Genome distance in bp to increase the size of bedpe rows. Default is 0 |
-h | --help | Help message |