Intersecting Loops
This code snippet shows how to intersect loops with intersect_bedpe in a Docker or Tinker environment.
We’ll start by assigning variables. First, our loops (called in Snippet: Calling Loops), then two .bed files, one containing CTCF sites and another with TSSs.
loops=my_loops.bedpeTSS=$HOME/lab-data/hg38/reference/GENCODE_TSSs_hg38.bedCTCF=$HOME/lab-data/hg38/reference/ENCODE3_cCRE-CTCF_hg38.bedOur output will contain loops that have at least one anchor that intersects with either a TSS or a CTCF site.
To print the intersecting loops in your terminal, use:
intersect_bedpe --bedpe $loops --bed_A $TSS --bed_B $CTCFTo save the intersecting loops to a file, use:
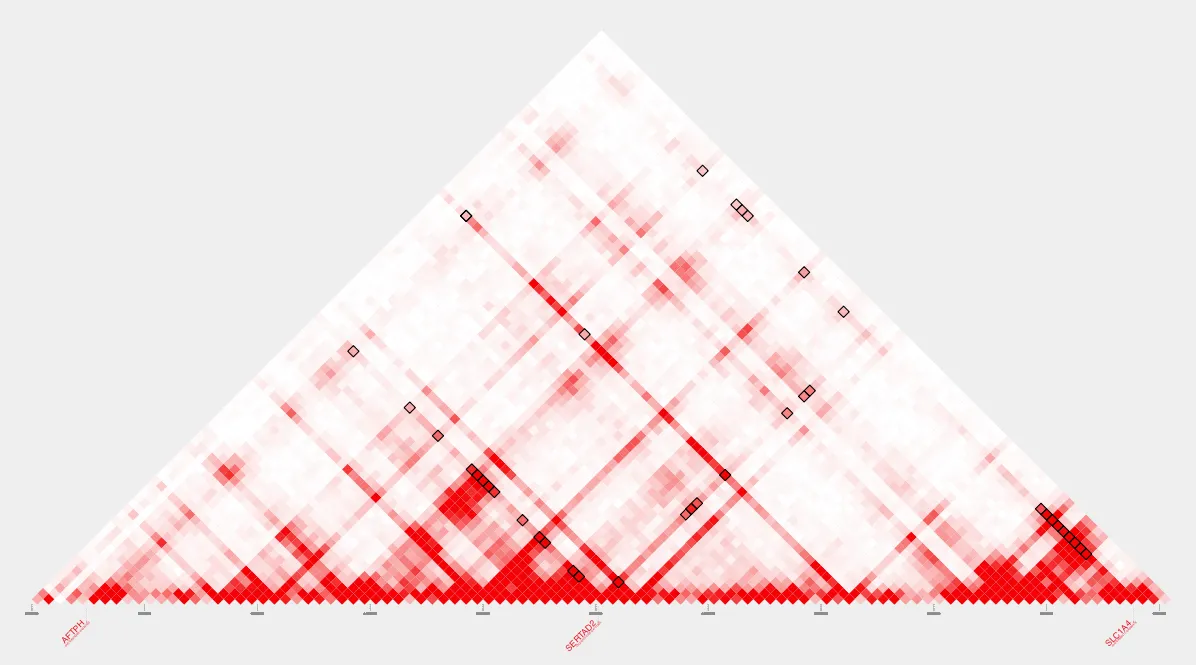
intersect_bedpe --bedpe $loops --bed_A $TSS --bed_B $CTCF > TSS-CTCF-loops.bedpeTo visualize the changes, let’s view our loops before and after using intersect_bedpe with plot_contacts.
Before intersecting with TSSs and CTCF sites:
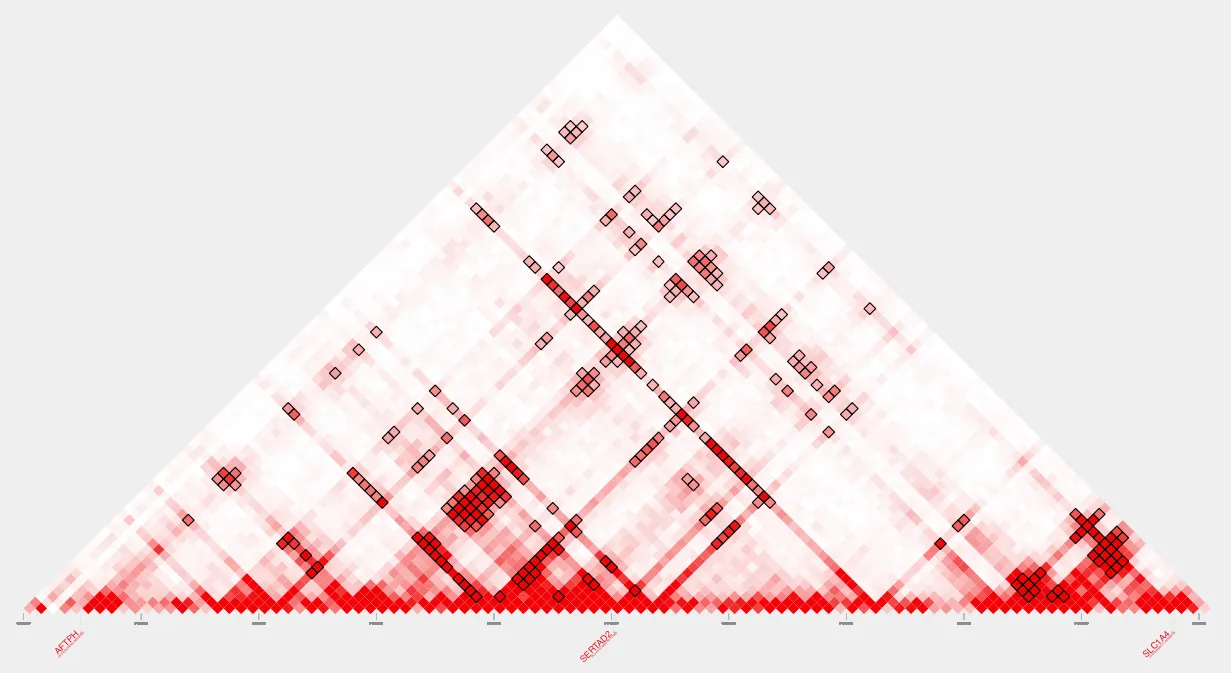
After intersecting with TSSs and CTCF sites: